Research Page for Ian Wilson
ian_j_wilson@outlook.com
Google Scholar Profile
This page contains links to example code, code fragments and source code that are used
in published and ongoing work.
Public Github repositories
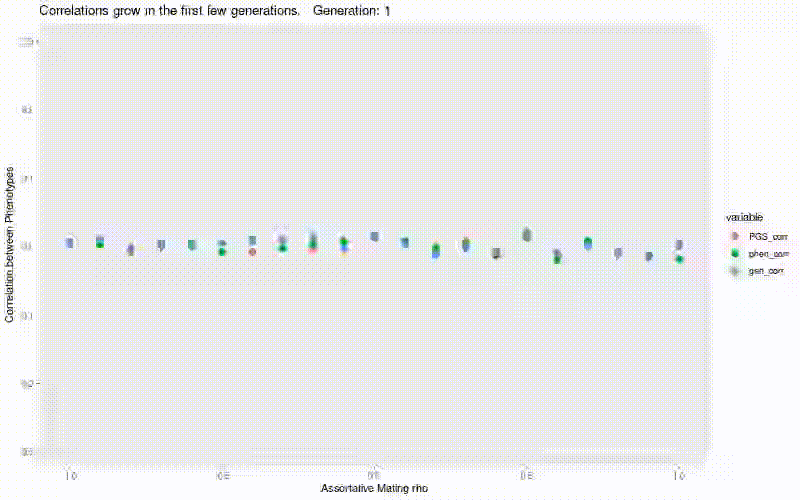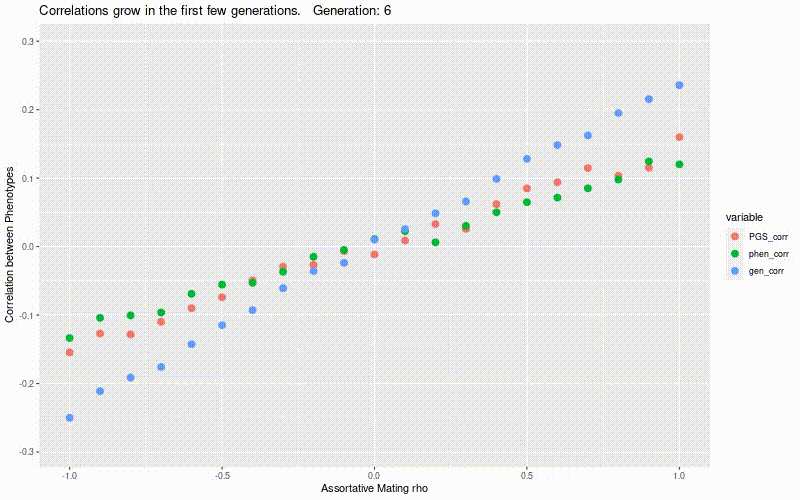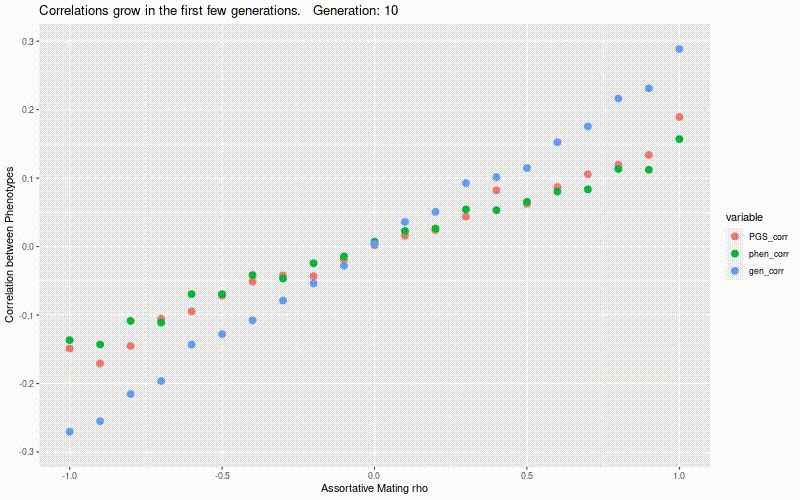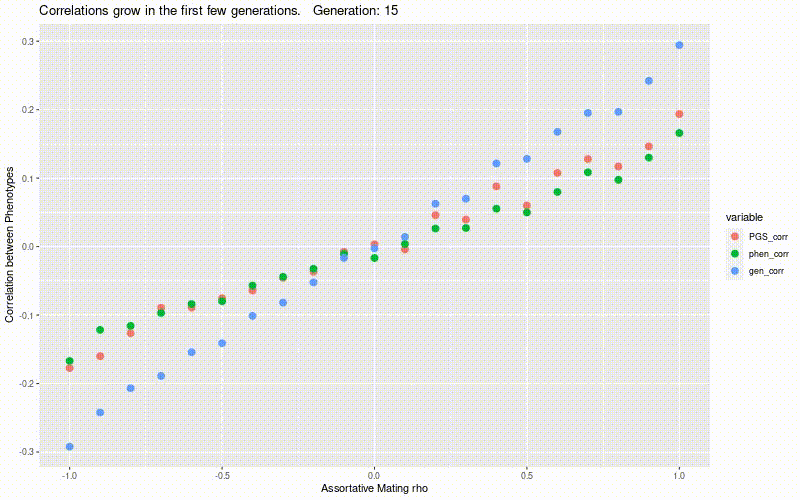
Simulate genetic correlation induced by assortative mating.
 Shiny applications for teaching population genetics
Shiny applications for teaching population genetics
- Bottleneck Selection
- Selection through a bottleneck
- SnakeDrift
- Simple Pictoral representation of Genetic Drift
- SelectionDriftSimulation
- Simulation of Drift and Selection
R packages
Packages (with varying degree of completeness) for the
R software system. Latest versions
in my R package directory
Install packages on unix/linux using the commands
R CMD INSTALL packagename.tar.gz
To install on a windows systems use the zip versions
and install from within R using
install.packages("packagename.zip",repos=NULL)
Where you have downloaded the package to your working directory. Once installed the
zip file can be safely deleted.
| genealogy.tar.gz |
genealogy.zip |
A package for the simulation of simple population genetic models |
| genomic.tar.gz |
genomic.zip |
A package for downloading and using HAPMAP and Perlegen
datasets |
| pac.tar.gz |
pac.zip |
A package using Li & Stephens (2003) model for the
analysis of genetic data, and my extensions for the analysis of
data from subdivided populations. |
| ARG.tar.gz |
ARG.zip |
Simulate and extract information from Ancestral Recombination
Graphs. Requires the ape package for plots. |
| batwing.tar.gz |
batwing.zip |
Package for postprocessing of BATWING output. |
| ijwtools.tar.gz |
ijwtools.zip |
My own set of utility functions (mainly for dealing with genetic data and the results from my own programs)
including quick reading from xml files, omnibus tests, Manhattan distances
between rows in a table, multivariate D' calculation. |
| snptree |
|
Create and plot lexical trees from SNP data. Used in paper
Recent mitochondrial DNA mutations increase the risk of developing multiple common late-onset human diseases,
by Hudson et al. link
|
Screenshots of interactive plots from the ARG package. Left plot
shows a view of an ancestral graph showing genetic variants (bottom
panel) along with the tree under a single site (top left panel) and
tree heights and lengths (top right panel). Right plot gives views
of trees 100 sites apart. Click on either plot for a larger
view.
C/C++ Software
Installation on Unix Systems
All Software in this section is in the form of zipped tar files.
Use
gunzip -c filename.tar.gz | tar xf -
then type make
to compile the program. A limited manual for micsat is in the file
micsat.doc. Other manuals can be downloaded in pdf format.
Windows (95/98/NT)
The programs are distributed as executable files in a zip file format
Macintosh (MacOS 8.0 or above)
A binhex-ed executable for the Macintosh, MacBatwing, may be downloaded.
On launching, the program opens a dialog box with a text field. The
user puts the program arguments (infile outfile seed) into this
field and hits ok.
(Compilation on the Mac courtesy of Oliver Pybus - send
comments/bugs to Ian.Wilson@ncl.ac.uk).
Program Descriptions
 |
Micsat
This is the program described in the paper "Genealogical
inference from microsatellite data" by Wilson and Balding published
in Genetics 150: 499-510.
A description of the program and
the downloaded files.
|
|
Bayesian Analysis of Trees With Internal
Node Generation. Batwing is described in
Wilson, Weale & Balding 2003.Inferences from DNA data:
population histories, evolutionary processes and forensic match
probabilities. Journal of the Royal Statistical Society: Series
A, 166: 155-188.
Download
|
 |
Parentage
Program described in Emery, Wilson, Craig, Boyle and Noble
(2001). Assignment of paternity groups without access to parental
genotypes: multiple mating and development plasticity in
squid.
Molecular Ecology: 10: 1265-1278.
download
The Computational Biology Service Unit at Cornell University
provides free world-wide access to parentage via a web interface (BioHPC.org).
Using this interface, researchers can run a variety of computationally
intensive bioinformatics and population genetics tools, Parentage being one of them.
Web inferface at BioHP.org
|
Hapgen
A program for haplotype reconstruction from genotypes. Work done with David
Cooper.
This program is a Markov chain Monte Carlo method to reconstruct
haplotypes from genotypes. It uses Metropolis coupled chains to
speed up mixing.
Includes options to deal with:
- Known phase,
- Missing Data,
- Multiple subpopulations
Ancestors
Inference of migration or population splitting using importance
sampling.
Vickers, McLeod, Spector & Wilson (2001). Assessment of mechanism
of skewed X-inactivation by analysis of twins.
BLOOD: 97:1274-1281
WinBUGS analysis of Twins
Multivariate Normal Model, and Twins Stem Cell Number
ian_j_wilson@outlook.com
Last Modified 18 June 2024






